Quick Start
Here’s a basic example of how to use midlearn to explain a trained LightGBM model, utilizing the familiar scikit-learn API.
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.metrics import root_mean_squared_error
from sklearn.datasets import fetch_openml
from sklearn import set_config
import lightgbm as lgb
import midlearn as mid
# Set up plotnine theme for clean visualizations
import plotnine as p9 # require plotnine >= 0.15.0
p9.theme_set(p9.theme_538(base_family='serif'))
# Configure scikit-learn display
set_config(display='text')
Error importing in API mode: ImportError('On Windows, cffi mode "ANY" is only "ABI".')
Trying to import in ABI mode.
1. Train a Black-Box Model
We use the California Housing dataset to train a LightGBM Regressor, which will serve as our black-box model.
# Load and prepare data
bikeshare = fetch_openml(data_id=42712)
X = pd.DataFrame(bikeshare.data, columns=bikeshare.feature_names)
y = bikeshare.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
# Fit a LightGBM regression model
estimator = lgb.LGBMRegressor(
force_col_wise=True,
n_estimators=500,
random_state=42
)
estimator.fit(X_train, y_train)
[LightGBM] [Info] Total Bins 283
[LightGBM] [Info] Number of data points in the train set: 13034, number of used features: 12
[LightGBM] [Info] Start training from score 190.379623
LGBMRegressor(force_col_wise=True, n_estimators=500, random_state=42)
model_pred = estimator.predict(X_test)
rmse = root_mean_squared_error(model_pred, y_test)
print(f"RMSE: {round(rmse, 6)}")
RMSE: 37.615267
2. Create an Explaination Model
We fit the MIDExplainer to the training data to create a globally faithful, interpretable surrogate model (MID).
# Initialize and fit the MID model
explainer = mid.MIDExplainer(
estimator=estimator,
penalty=.05,
singular_ok=True,
interactions=True,
encoding_frames={'hour':list(range(24))}
)
explainer.fit(X_train)
Generating predictions from the estimator...
R callback write-console: singular fit encountered
MIDExplainer(encoding_frames={'hour': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23]},
estimator=LGBMRegressor(force_col_wise=True, n_estimators=500,
random_state=42),
penalty=0.05, singular_ok=True)
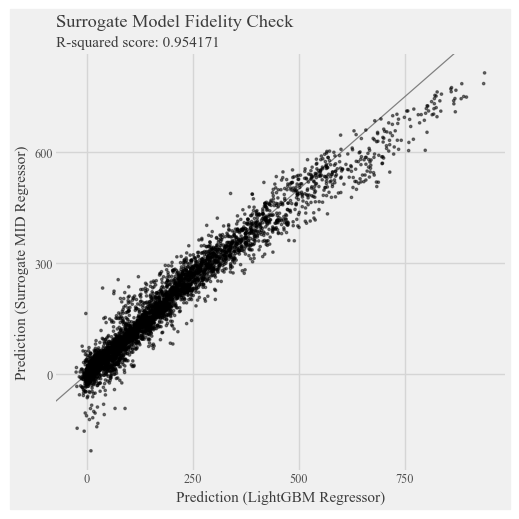
# Check the fidelity of the surrogate model to the original model
p = p9.ggplot() \
+ p9.geom_abline(slope=1, color='gray') \
+ p9.geom_point(p9.aes(estimator.predict(X_test), explainer.predict(X_test)), alpha=0.5, shape=".") \
+ p9.labs(
x='Prediction (LightGBM Regressor)',
y='Prediction (Surrogate MID Regressor)',
title='Surrogate Model Fidelity Check',
subtitle=f'R-squared score: {round(explainer.fidelity_score(X_test), 6)}',
)
display(p + p9.theme(figure_size=(5,5)))
Generating predictions from the estimator...
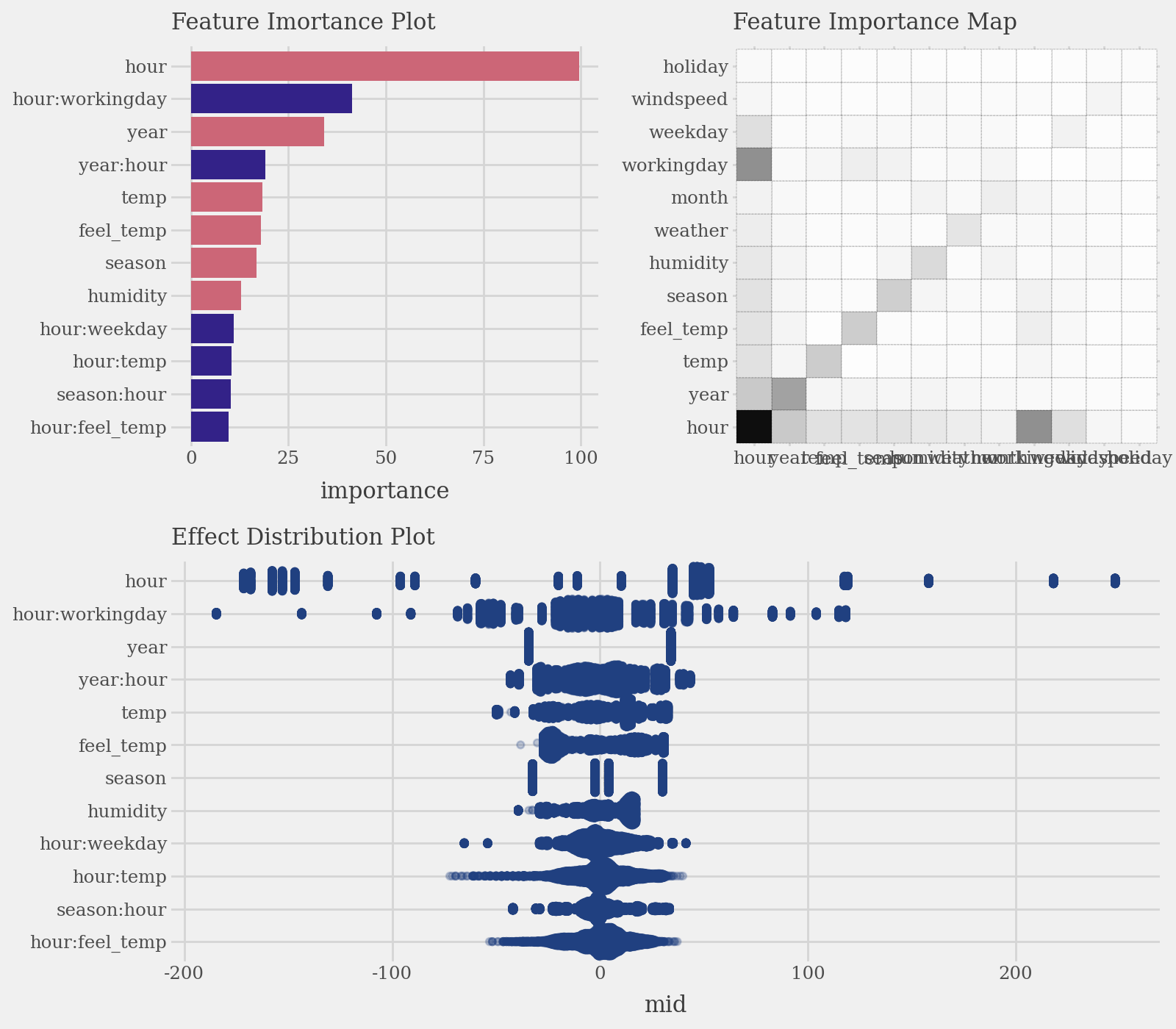
3. Visualize the Explanation Model
The MID model allows for clear visualization of feature importance, individual effects, and local prediction breakdowns.
# Calculate and plot overall feature importance (default bar plot and heatmap)
imp = explainer.importance()
p1 = (
imp.plot(max_nterms=12, theme = 'muted') +
p9.labs(subtitle="Feature Imortance Plot") +
p9.coord_flip()
)
p2 = (
imp.plot(style='heatmap', color='black', linetype='dotted') +
p9.labs(subtitle="Feature Importance Map")
)
p3 = (
imp.plot(max_nterms=12, color="#204080", alpha = .25, style='sina') +
p9.labs(subtitle="Effect Distribution Plot")
)
display(((p1 | p2) / p3) & p9.theme(figure_size=(8, 7), legend_position="none"))
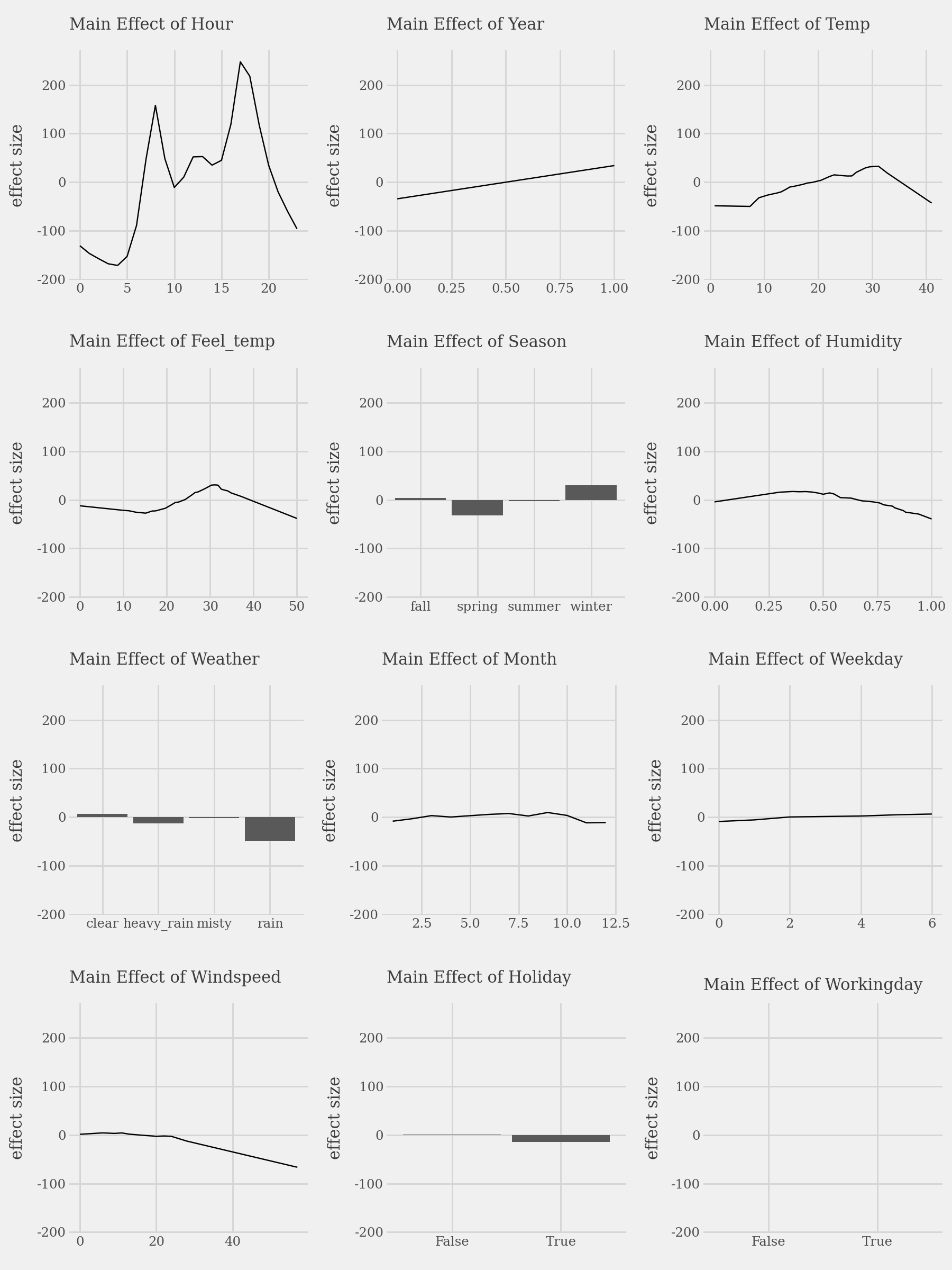
# Plot the top 3 important main effects (Component Functions)
plots = list()
for i, t in enumerate(imp.terms(interactions=False)):
p = (
explainer.plot(term=t) +
p9.lims(y=[-180, 250]) +
p9.labs(
subtitle=f"Main Effect of {t.capitalize()}",
x="",
y="effect size"
)
)
plots.append(p)
p1 = (
(plots[0] | plots[1] | plots[2]) /
(plots[3] | plots[4] | plots[5]) /
(plots[6] | plots[7] | plots[8]) /
(plots[9] | plots[10] | plots[11])
)
display(p1 + p9.theme(figure_size=(9, 12)))
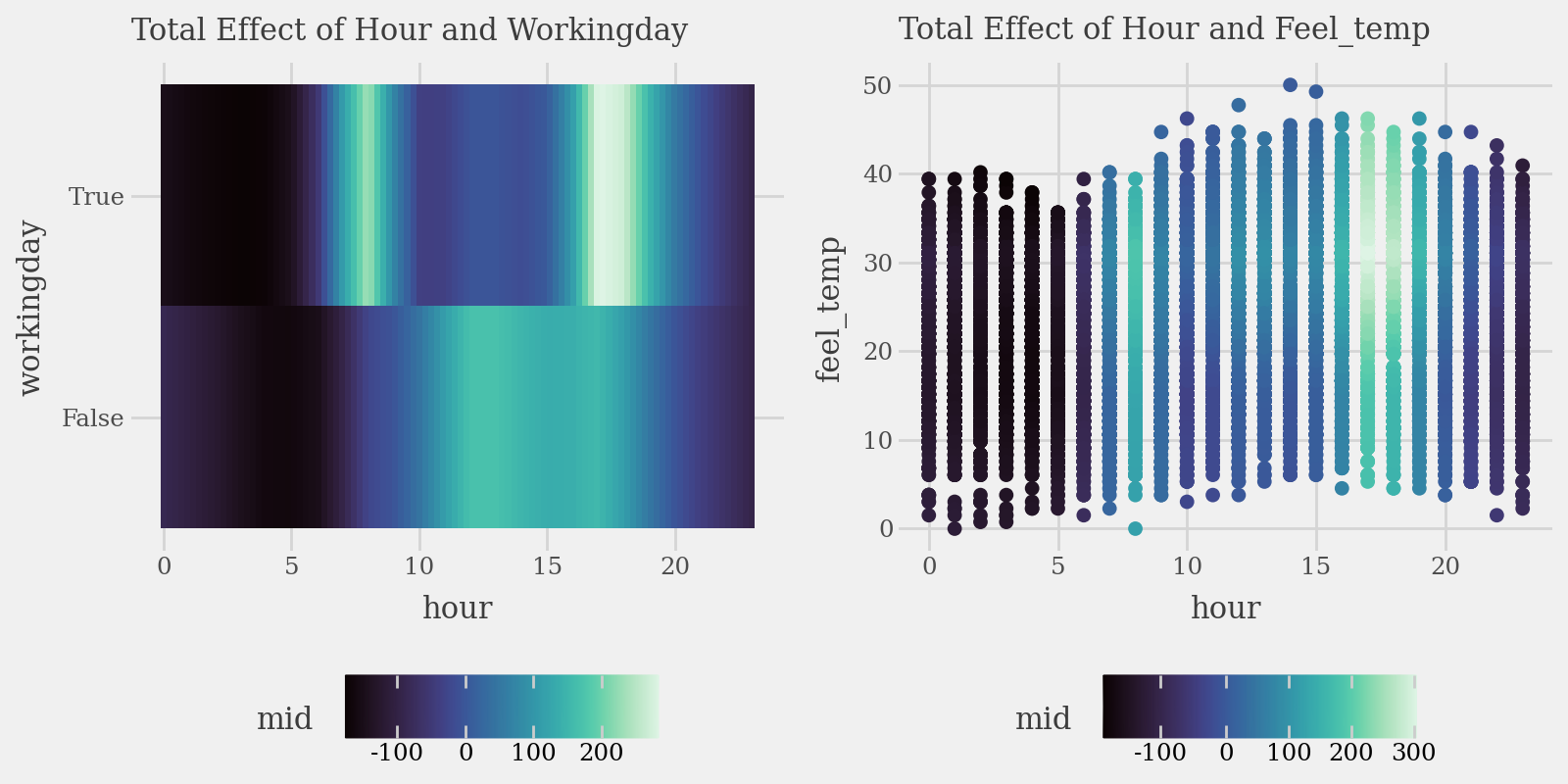
# Plot the interaction of pairs of variables (Component Functions)
p1 = (
explainer.plot(
"hour:workingday",
theme='mako',
main_effects=True
) +
p9.labs(subtitle="Total Effect of Hour and Workingday")
)
p2 = (
explainer.plot(
"hour:feel_temp",
style='data',
theme='mako',
data=X_train,
main_effects=True,
size=2
) +
p9.labs(subtitle="Total Effect of Hour and Feel_temp")
)
display((p1 | p2) & p9.theme(figure_size=(8, 4), legend_position="bottom"))
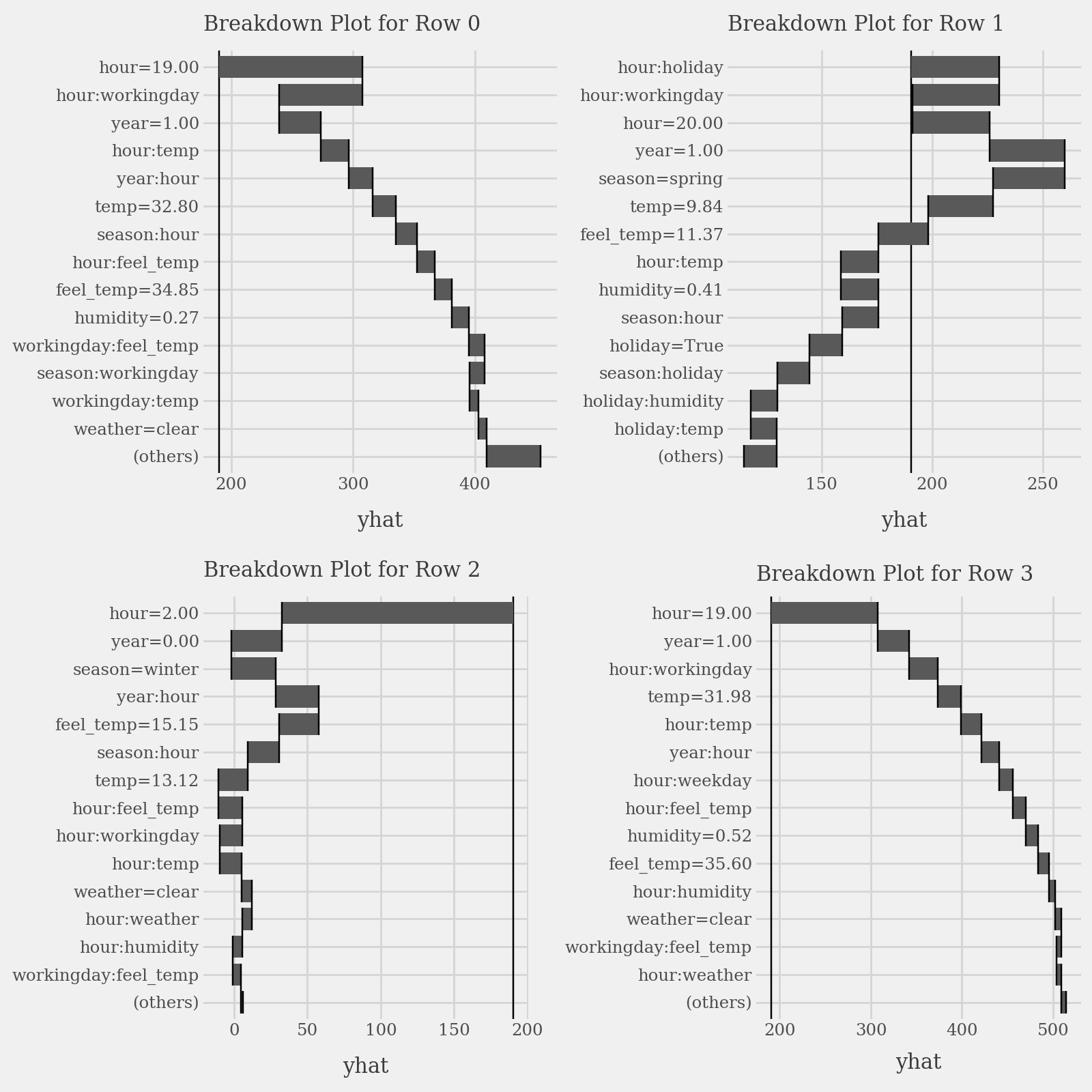
# Plot prediction breakdowns for the first three test samples (Local Interpretability)
plots = list()
for i in range(4):
p = (
explainer.breakdown(row=i, data=X_test).plot(format_args = {'digits': 2}) +
p9.labs(subtitle=f"Breakdown Plot for Row {i}")
)
plots.append(p)
p1 = (
(plots[0] | plots[1]) /
(plots[2] | plots[3])
)
display(p1 + p9.theme(figure_size=(8, 8)))
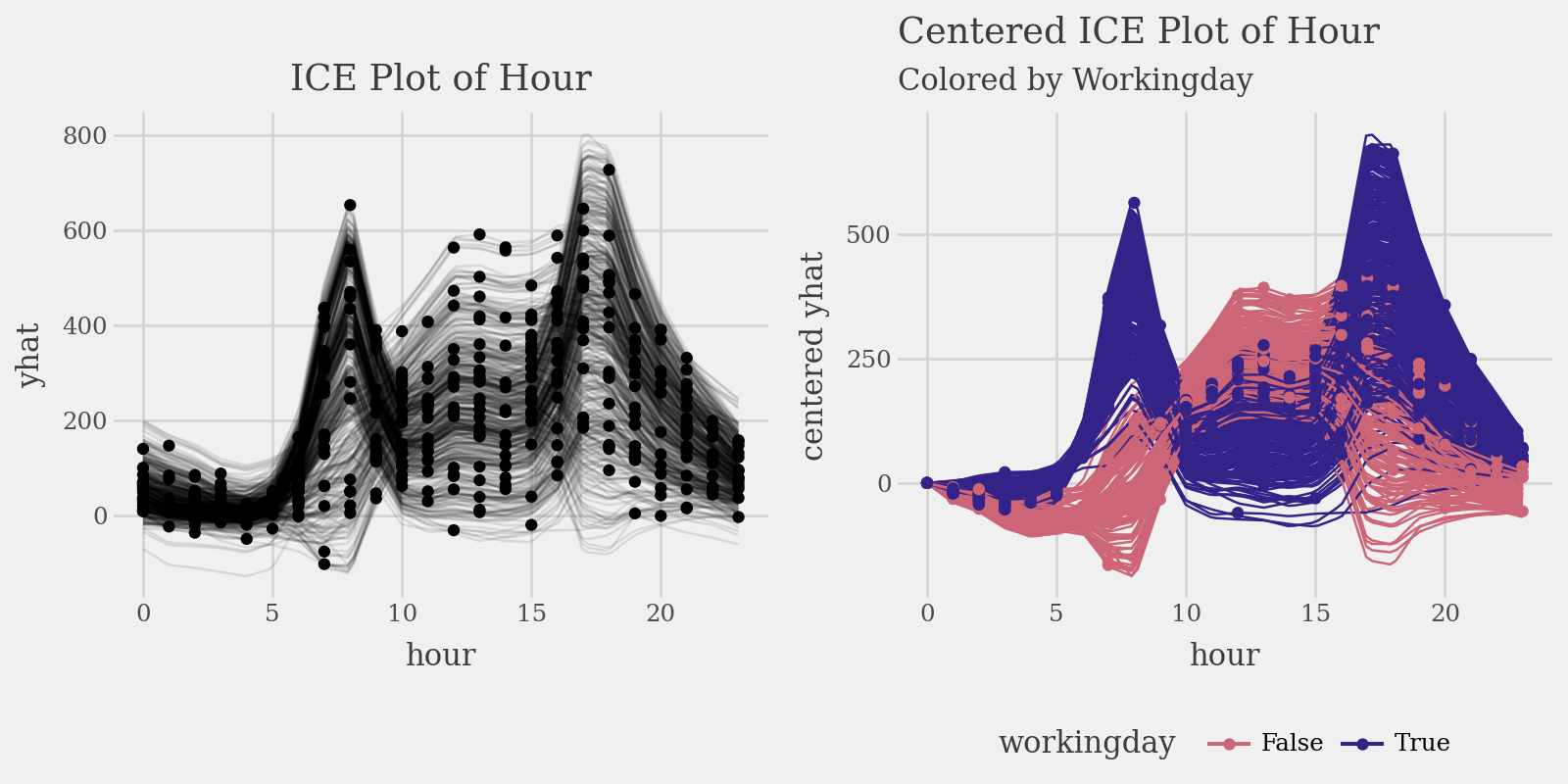
# Plot individual conditional expectations (ICE) with color encoding
ice = explainer.conditional(
variable='hour',
data=X_train.head(500)
)
p1 = (
ice.plot(alpha=.1) +
p9.ggtitle("ICE Plot of Hour")
)
p2 = (
ice.plot(
style='centered',
var_color='workingday',
theme='muted'
) +
p9.labs(
title="Centered ICE Plot of Hour",
subtitle="Colored by Workingday"
) +
p9.theme(legend_position="bottom")
)
display((p1 | p2) & p9.theme(figure_size=(8, 4), legend_position="bottom"))