Interpretation of Classification Models
Source:vignettes/articles/classification.Rmd
classification.RmdThis article presents some examples of the interpretation of
classification models using midr.
# load required packages
library(midr)
library(ggplot2)
library(gridExtra)
library(Metrics)
theme_set(theme_midr())Classification Task
We use the titanic dataset, which is available on the website https://www.encyclopedia-titanica.org/ and is included
in the DALEX package. The dataset has 9 variables for 2207
people, of which 1317 were passengers and 890 were crew members. We fit
some classification models that predict who survived the tragedy and who
did not, and then we interpret the fitted models.
# benchmark classification task
library(DALEX)
#> Welcome to DALEX (version: 2.5.3).
#> Find examples and detailed introduction at: http://ema.drwhy.ai/
#> Additional features will be available after installation of: ggpubr.
#> Use 'install_dependencies()' to get all suggested dependencies
set.seed(42)
test_rows <- sample(nrow(titanic), 500L)
train <- titanic[-test_rows, -5]
str(train)
#> 'data.frame': 1707 obs. of 8 variables:
#> $ gender : Factor w/ 2 levels "female","male": 2 1 1 2 2 1 2 2 2 2 ...
#> $ age : num 42 39 16 25 30 28 27 20 30 27 ...
#> $ class : Factor w/ 7 levels "1st","2nd","3rd",..: 3 3 3 3 2 2 3 3 3 3 ...
#> $ embarked: Factor w/ 4 levels "Belfast","Cherbourg",..: 4 4 4 4 2 2 2 4 4 4 ...
#> $ fare : num 7.11 20.05 7.13 7.13 24 ...
#> $ sibsp : num 0 1 0 0 1 1 0 0 0 0 ...
#> $ parch : num 0 1 0 0 0 0 0 0 0 0 ...
#> $ survived: Factor w/ 2 levels "no","yes": 1 2 2 2 1 2 2 2 1 1 ...
test <- titanic[ test_rows, -5]
str(test[, -9])
#> 'data.frame': 500 obs. of 8 variables:
#> $ gender : Factor w/ 2 levels "female","male": 2 2 1 2 2 2 2 2 1 2 ...
#> $ age : num 74 19 32 21 40 23 24 26 34 28 ...
#> $ class : Factor w/ 7 levels "1st","2nd","3rd",..: 3 3 2 3 4 3 5 5 2 5 ...
#> $ embarked: Factor w/ 4 levels "Belfast","Cherbourg",..: 4 2 4 4 4 4 4 4 4 1 ...
#> $ fare : num 7.15 7.04 21 8.08 0 ...
#> $ sibsp : num 0 0 0 0 0 0 0 0 1 0 ...
#> $ parch : num 0 0 0 0 0 0 0 0 1 0 ...
#> $ survived: Factor w/ 2 levels "no","yes": 1 1 2 1 1 1 2 1 2 2 ...For each model type, we fit a classification model using the training dataset of 1707 people and an interpretative MID surrogate of the target model using the same dataset. We then evaluate the predictive accuracy of the target model by AUC and the representation accuracy of the surrogate model by the Spearman’s rank correlation coefficient between two predicted probabilities.
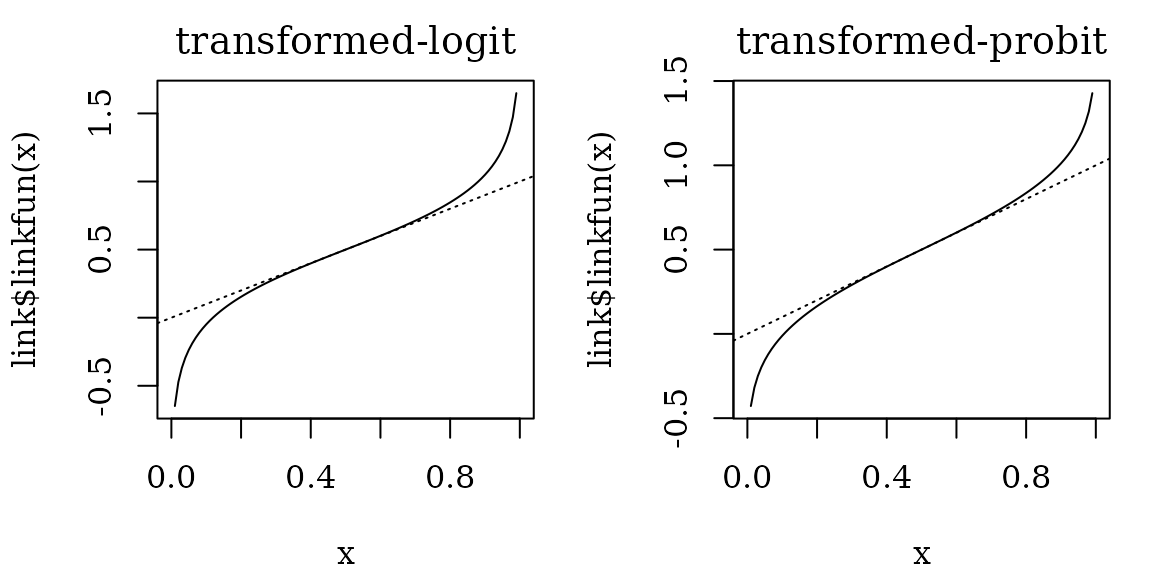
In the following examples, we use two specialized link functions for
classification tasks: translogit (transformed-logit) and
transprobit (transformed-probit). These two link functions
are transformed so that
and
.
With these link functions, the effects on the linear predictor can be
approximately interpreted as the upper bound of the effects on the
predicted probabilities.
# define utility functions for the following chunks
effect_plots <- function(object) {
plots <- mid.plots(mid, terms = mid.terms(mid)[1:6])
for (i in 1:6) {
plots[[i]] <- plots[[i]] + ggtitle("main effect")
if (any(i == c(1, 3, 4)))
plots[[i]] <- plots[[i]] + coord_flip()
}
plots
}
interaction_plot <- function(
object, term = NULL, theme = "shap") {
if (is.null(term))
term <- mid.terms(mid.importance(object), main.effect = FALSE)[1L]
ggmid(object, term, type = "data", data = na.omit(titanic),
theme = theme, main.effects = TRUE) +
theme(legend.position = "right") +
ggtitle("interaction effect plot")
}
ice_plot <- function(object, variable = "age") {
ggmid(mid.conditional(object, variable,
data = na.omit(titanic)[1:200, ]),
var.color = gender, theme = "shap_r") +
theme(legend.position = "right") +
ggtitle("conditional expectation")
}
importance_plot <- function(object) {
ggmid(mid.importance(object), "heatmap", theme = "grayscale") +
theme(legend.position = "right") +
ggtitle("feature importance")
}
evaluation_plot <- function(model, mid, ...) {
pred <- get.yhat(model, test, ...)
pred_mid <- get.yhat(mid, test)
actual <- as.numeric(test$survived == "yes")
auc_vs_test <- auc(actual, pred)
cor_vs_mid <- cor(pred, pred_mid, method = "spearman",
use = "pairwise.complete.obs")
ggplot() + scale_color_theme("Accent") +
geom_point(aes(x = pred, y = pred_mid), col = "#4378bf",
data = na.omit(data.frame(pred, pred_mid))) +
geom_abline(slope = 1, intercept = 0, col = "black", lty = 2) +
theme(legend.position = "right") + xlim(0, 1) +
labs(x = "model-prediction", y = "mid-prediction") +
annotate("text", family = "serif", size = 3, x = 0.2, y = 0.8,
label = sprintf("vs test (AUC) : %.3f\nvs mid (Spearman): %.3f",
auc_vs_test, cor_vs_mid)
) + ggtitle("representation accuracy")
}Additive Models
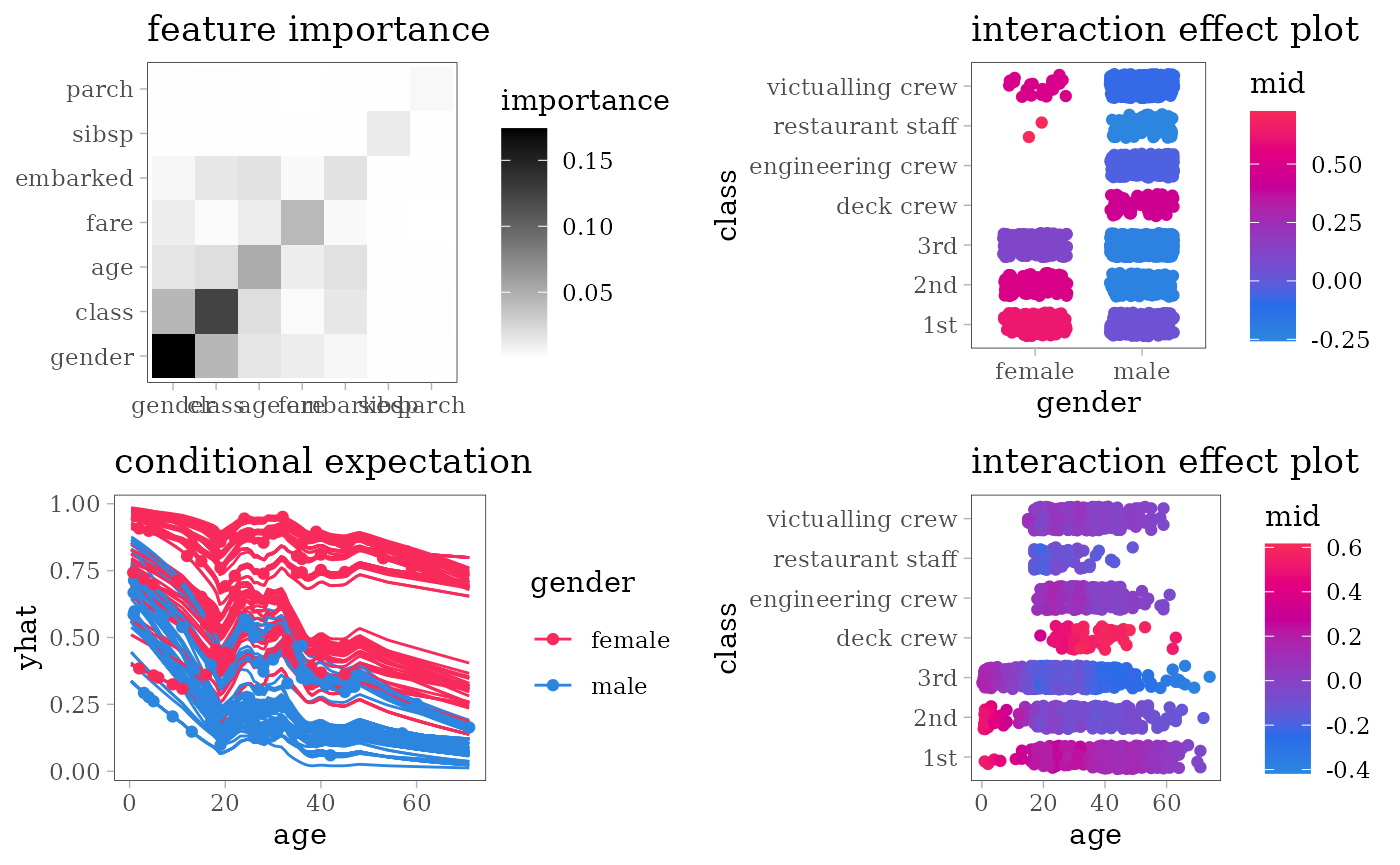
Logistic Regression
model <- glm(survived == "yes" ~ ., family = "binomial", data = train)
mid <- interpret(survived ~ .^2, train, model, link = "translogit")
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, model = model,
#> link = "translogit")
#>
#> Model Class: glm, lm
#>
#> Intercept: 0.26975
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0
grid.arrange(grobs = effect_plots(mid), nrow = 2L)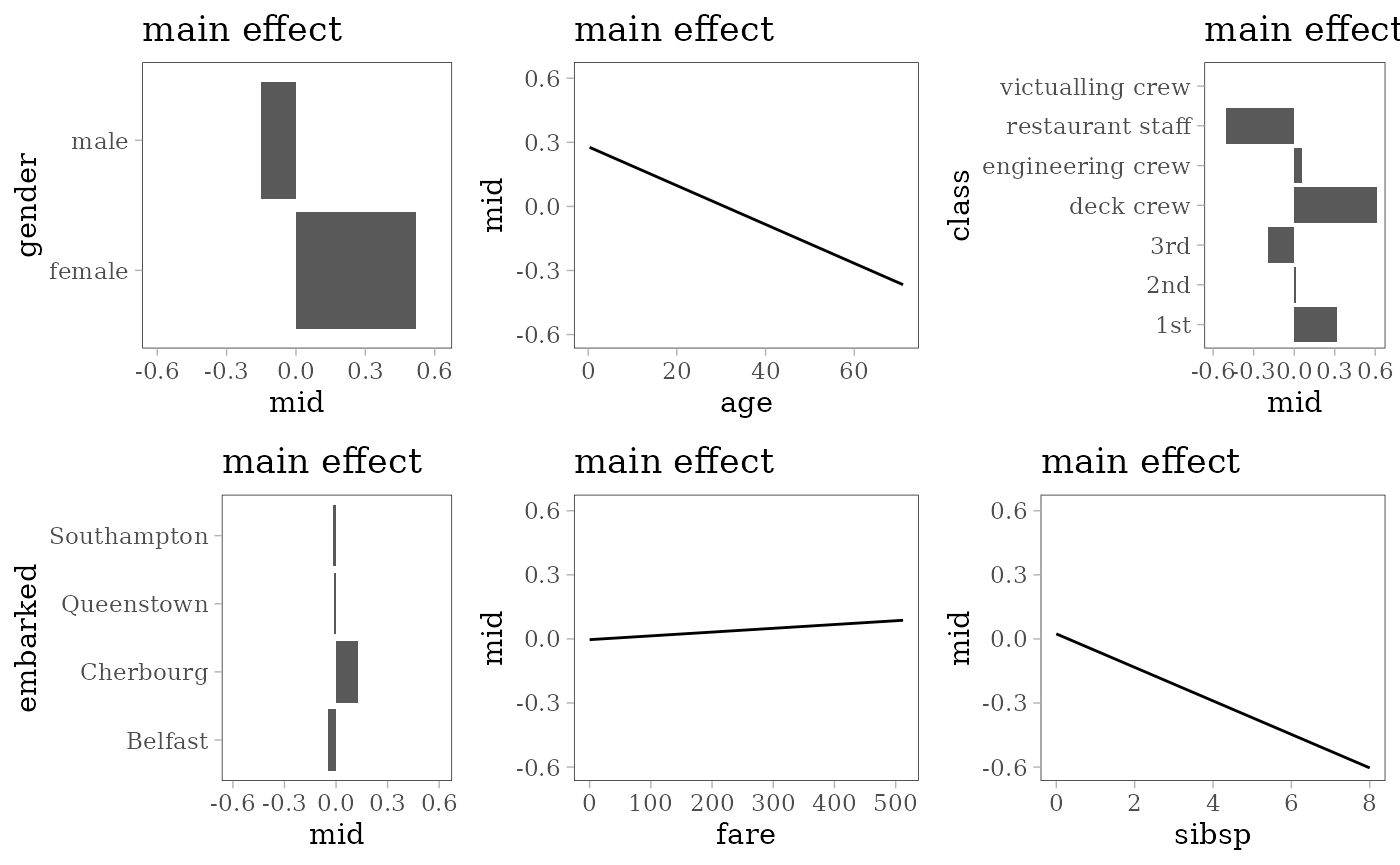
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
evaluation_plot(model, mid, target = "yes"))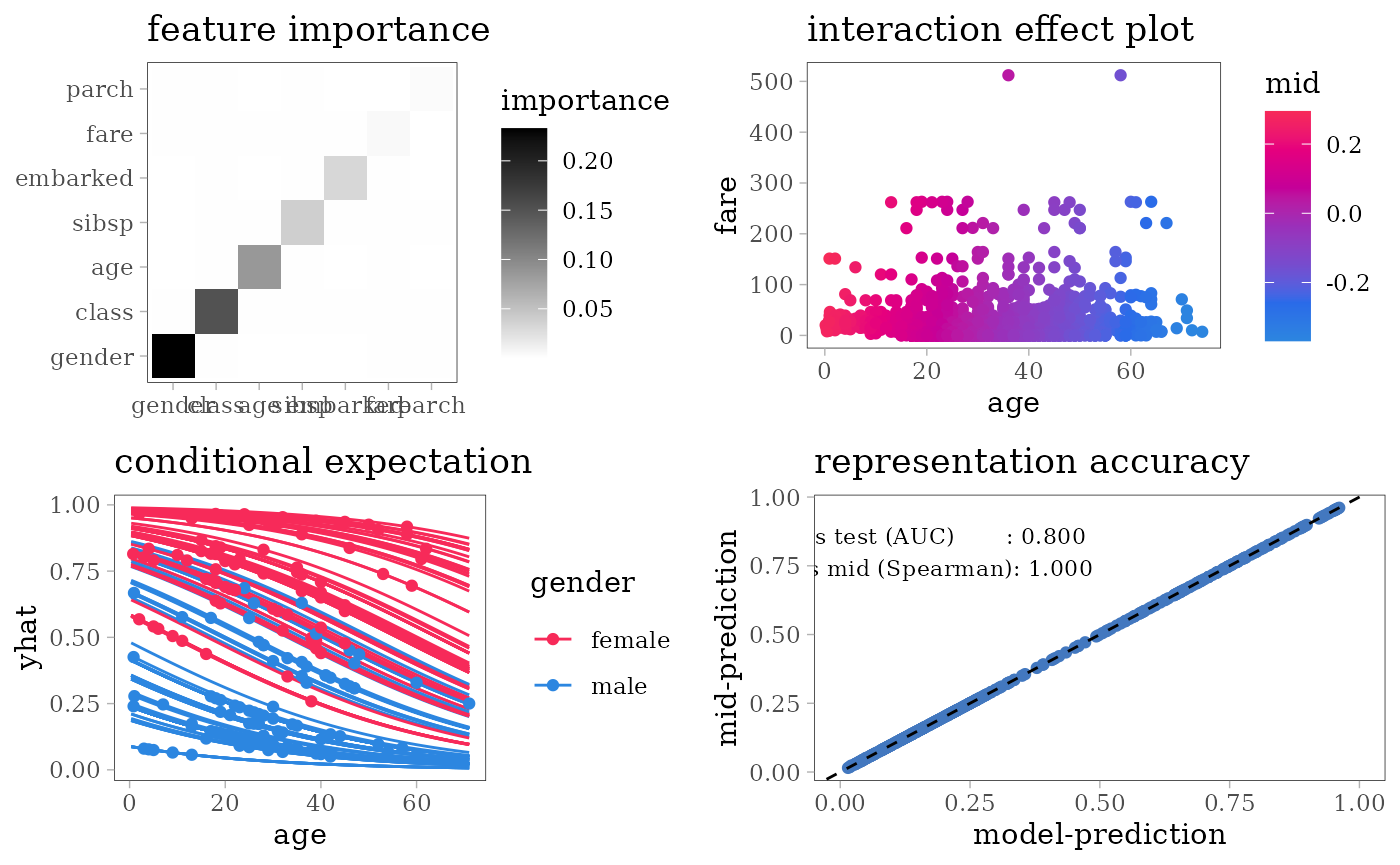
Neural Network
Single Hidden Layer Network
library(nnet)
set.seed(42)
model <- nnet(survived ~ ., train, size = 5, maxit = 1e3, trace = FALSE)
mid <- interpret(survived ~ .^2, train, model, link = "transprobit",
lambda = .01)
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, model = model,
#> link = "transprobit", lambda = 0.01)
#>
#> Model Class: nnet.formula, nnet
#>
#> Intercept: 0.29558
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0.045592
grid.arrange(grobs = effect_plots(mid), nrow = 2L)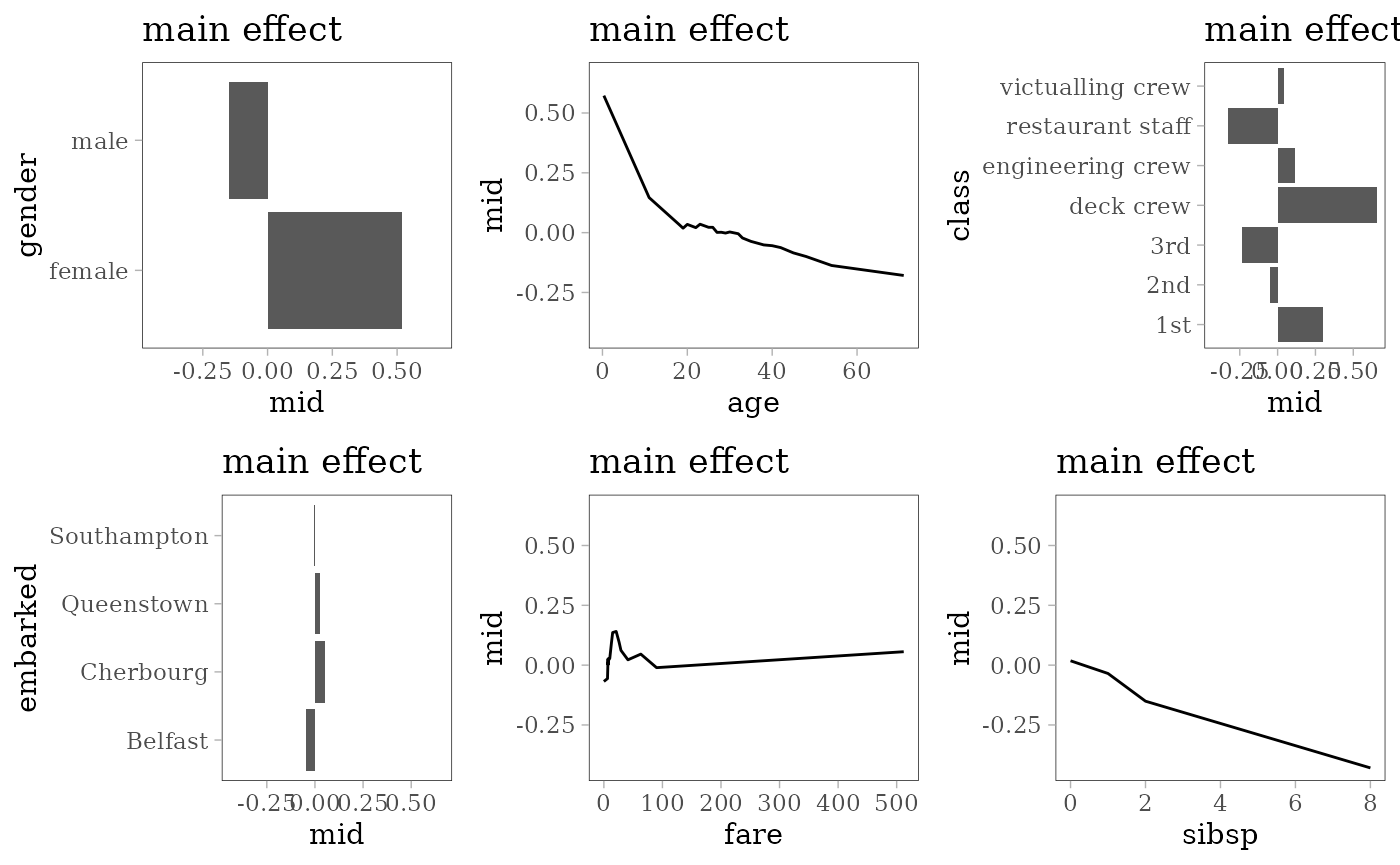
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
evaluation_plot(model, mid))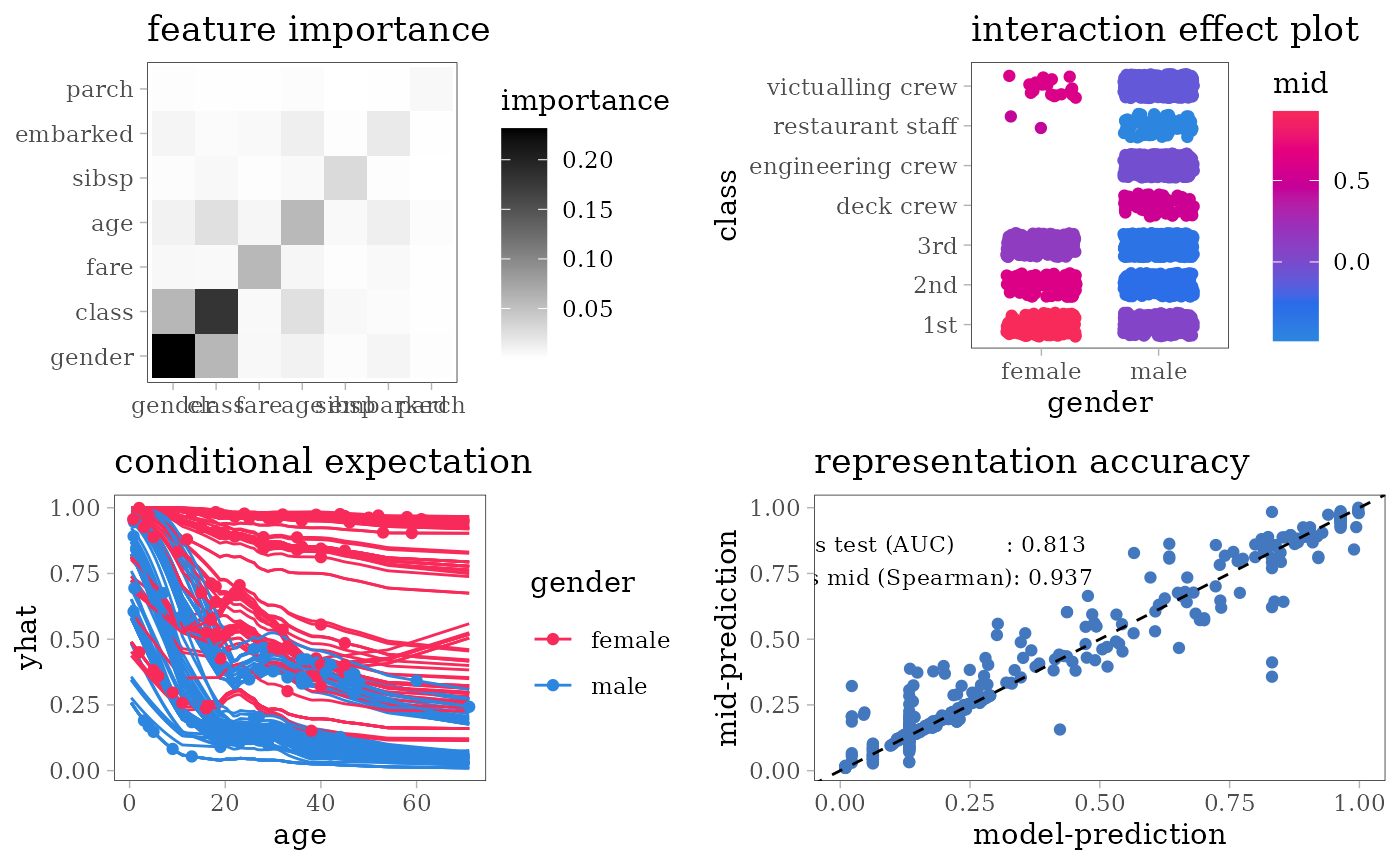
Support Vector Machine
RBF Kernel SVM
library(e1071)
#>
#> Attaching package: 'e1071'
#> The following object is masked from 'package:ggplot2':
#>
#> element
model <- svm(survived ~ ., train, kernel = "radial", probability = TRUE)
mid <- interpret(survived ~ .^2, train, model, link = "transprobit",
pred.args = list(target = "yes"))
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, model = model,
#> pred.args = list(target = "yes"), link = "transprobit")
#>
#> Model Class: svm.formula, svm
#>
#> Intercept: 0.29569
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0.0073507
grid.arrange(grobs = effect_plots(mid), nrow = 2L)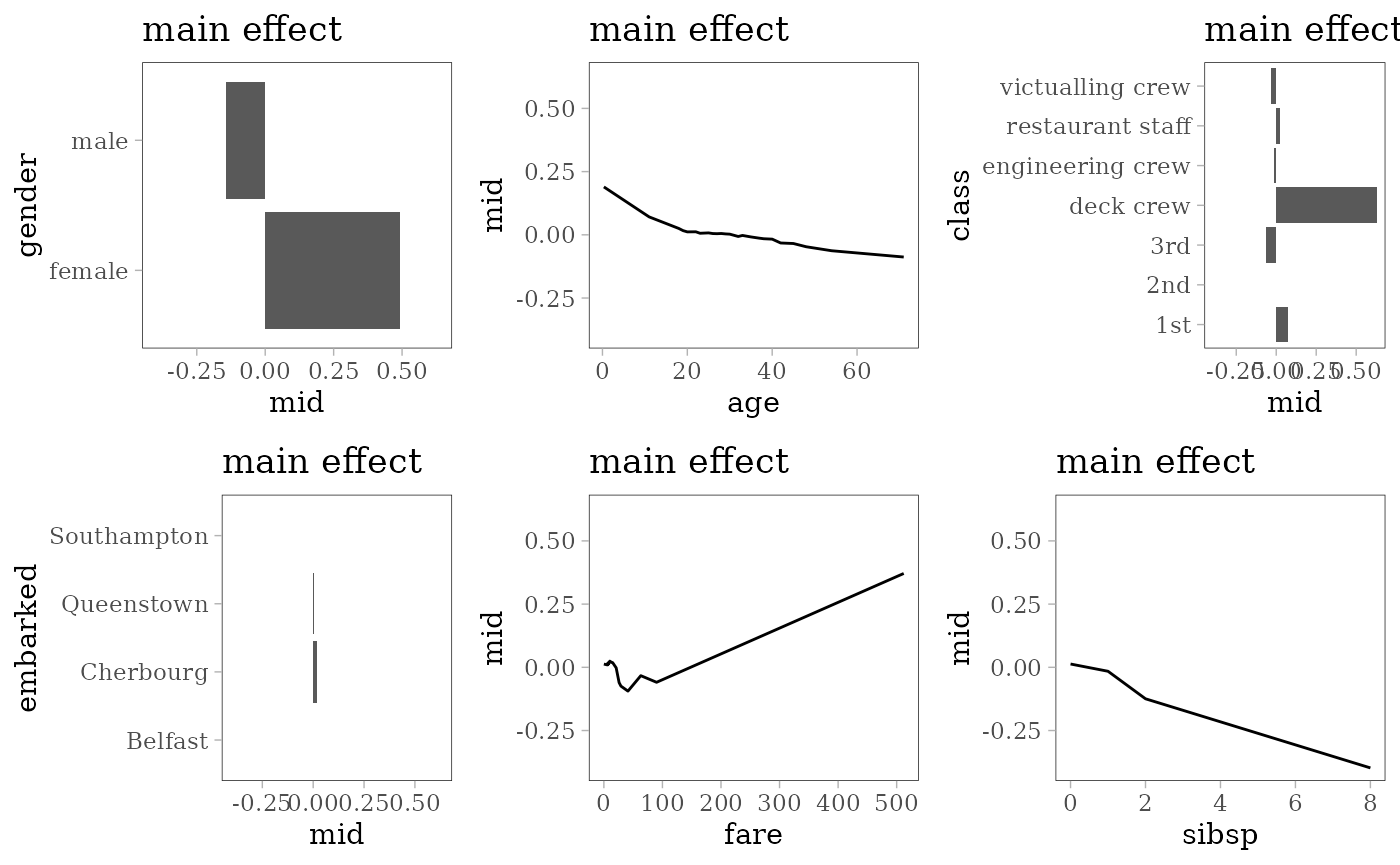
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
evaluation_plot(model, mid, target = "yes"))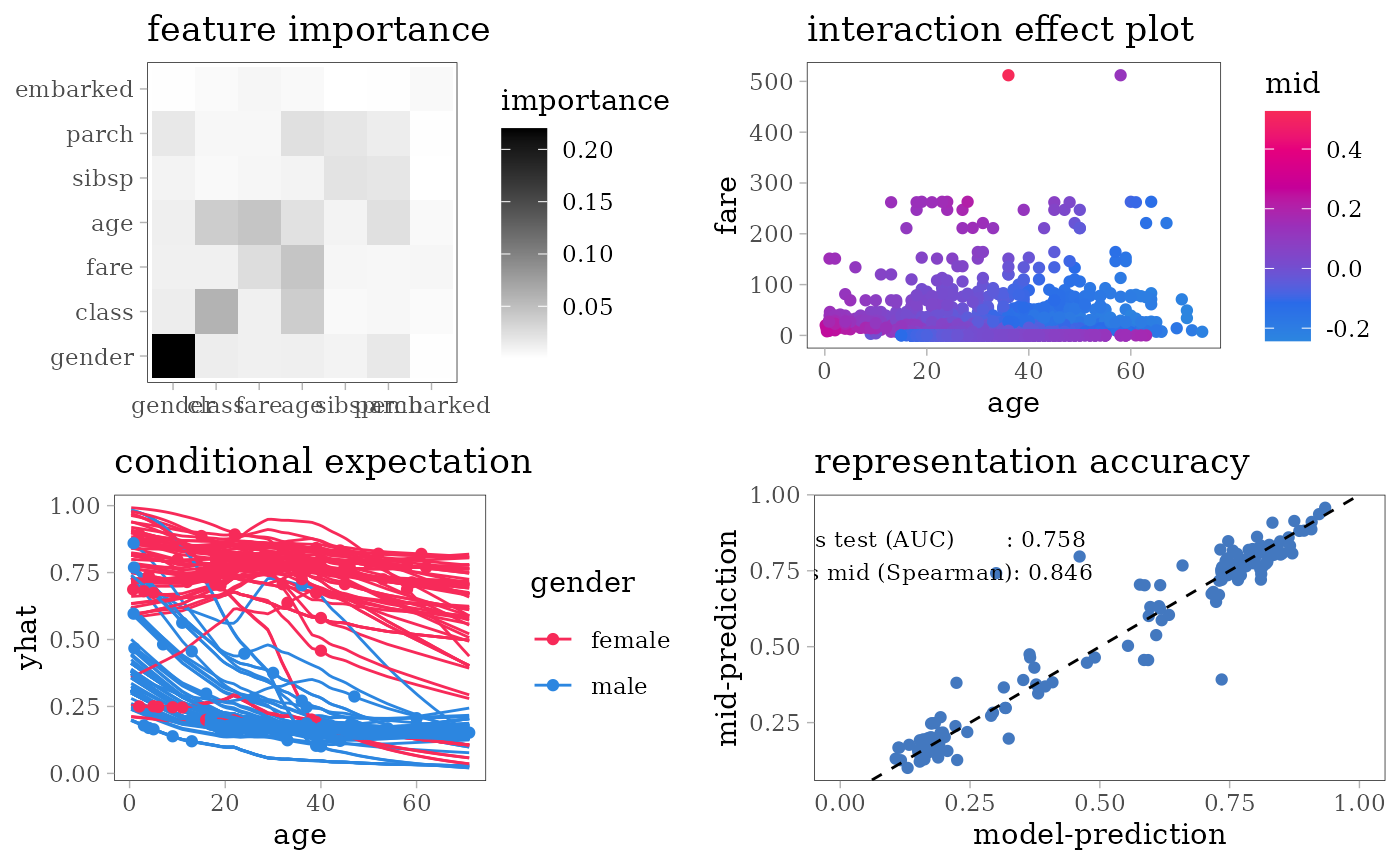
Tree Based Models
Random Forest
library(ranger)
set.seed(42)
model <- ranger(survived ~ ., na.omit(train), probability = TRUE)
mid <- interpret(survived ~ .^2, train, model,
link = "transprobit", lambda = .01)
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, model = model,
#> link = "transprobit", lambda = 0.01)
#>
#> Model Class: ranger
#>
#> Intercept: 0.30031
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0.062185
grid.arrange(grobs = effect_plots(mid), nrow = 2L)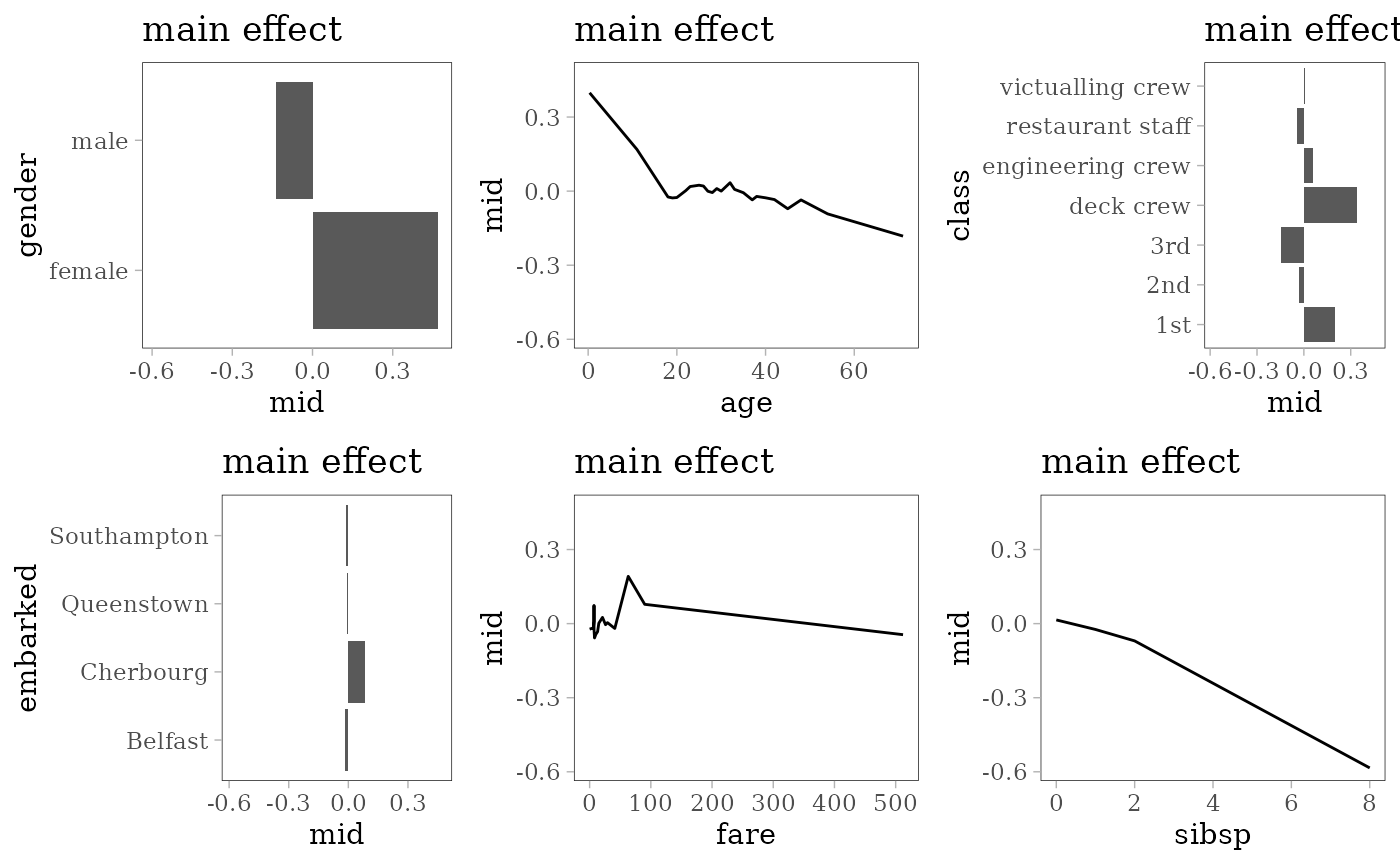
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
evaluation_plot(model, mid, target = "yes"))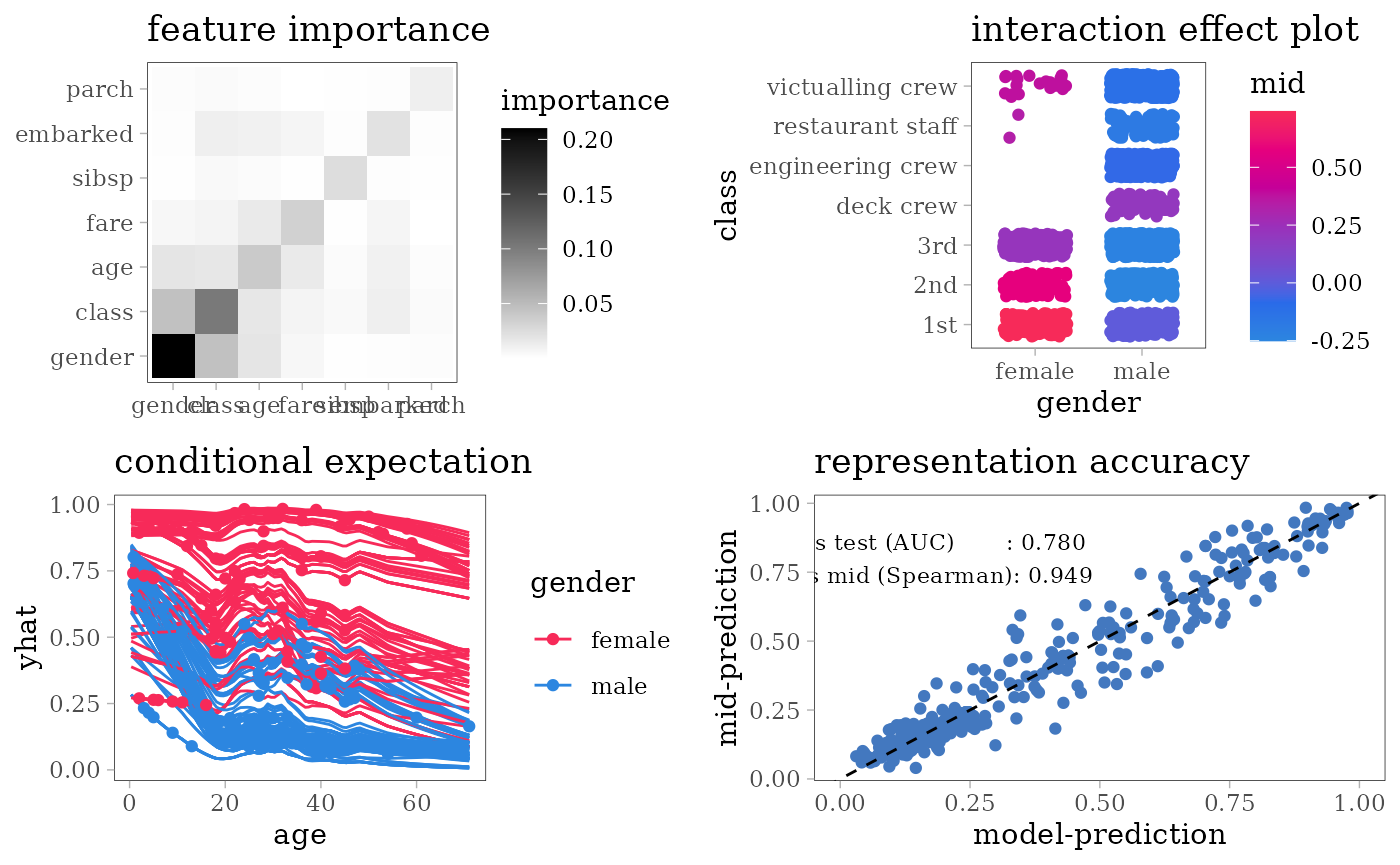
Decision Tree
library(rpart)
model <- rpart(survived ~ ., train)
# create encoding frames for CART
frm <- cbind(model$frame, labels(model, collapse = FALSE))
print(t(frm[frm$var != "<leaf>", c("var", "ltemp")]))
#> 1 2 4 9 5 11 3 6
#> var "gender" "class" "age" "sibsp" "age" "fare" "class" "fare"
#> ltemp "b" "bcefg" ">=9.5" ">=2.5" ">=54.5" ">=26.63" "c" ">=24.56"
#> 13
#> var "age"
#> ltemp ">=36.5"
fun <- function(x) if (is.numeric(x)) range(x, na.rm = TRUE) else levels(x)
frames <- lapply(train, fun)
frames$age <- c(frames$age, 9.5, 54.5, 36.5)
frames$fare <- c(frames$fare, 26.63, 24.56)
frames$sibsp <- c(frames$fare, 2.5)
mid <- interpret(survived ~ .^2, train, model, link = "transprobit",
singular.ok = TRUE, type = 0, frames = frames)
#> singular fit encountered
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, model = model,
#> link = "transprobit", singular.ok = TRUE, type = 0, frames = frames)
#>
#> Model Class: rpart
#>
#> Intercept: 0.29099
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0.022918
grid.arrange(grobs = effect_plots(mid), nrow = 2L)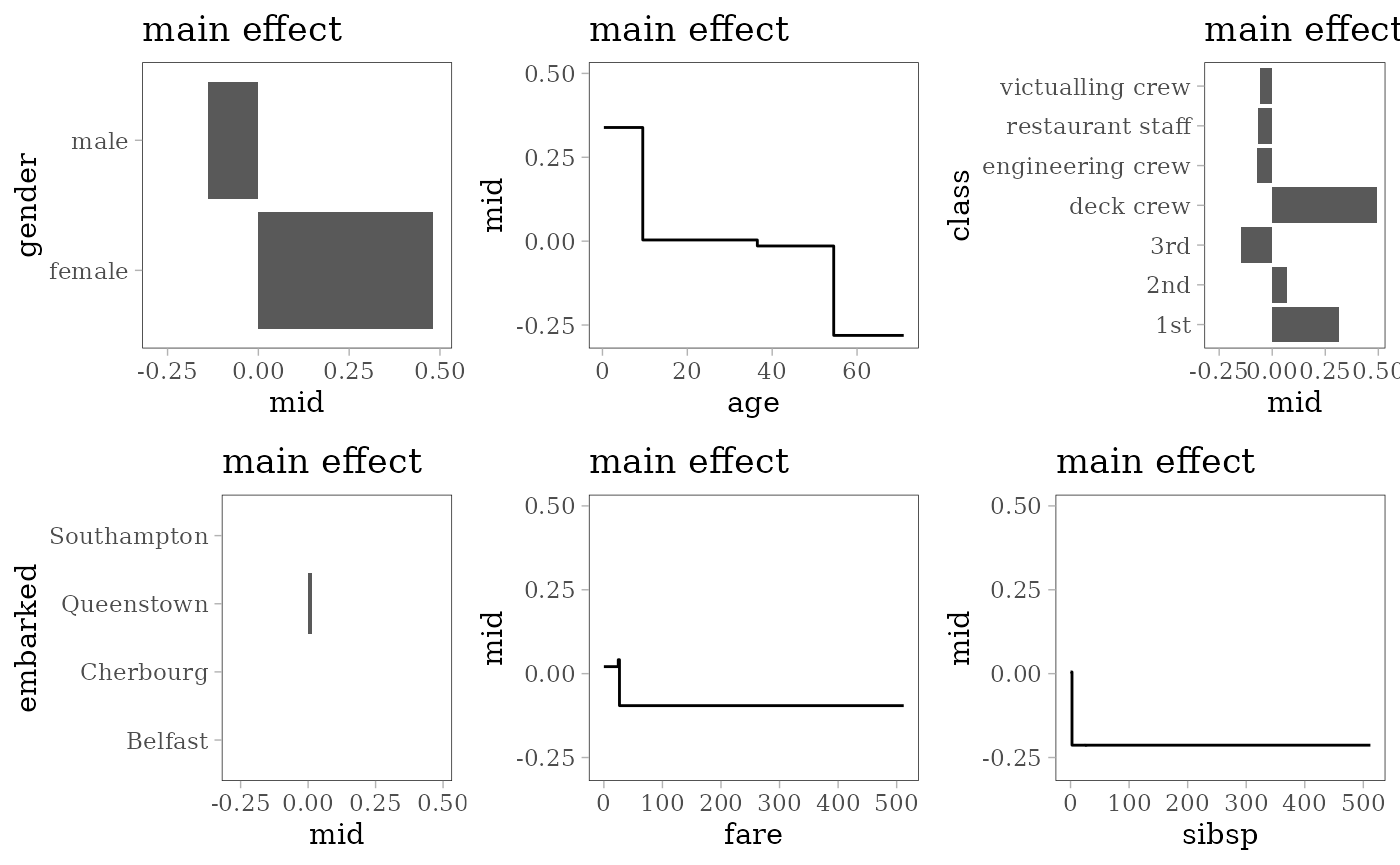
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
evaluation_plot(model, mid, target = "yes"))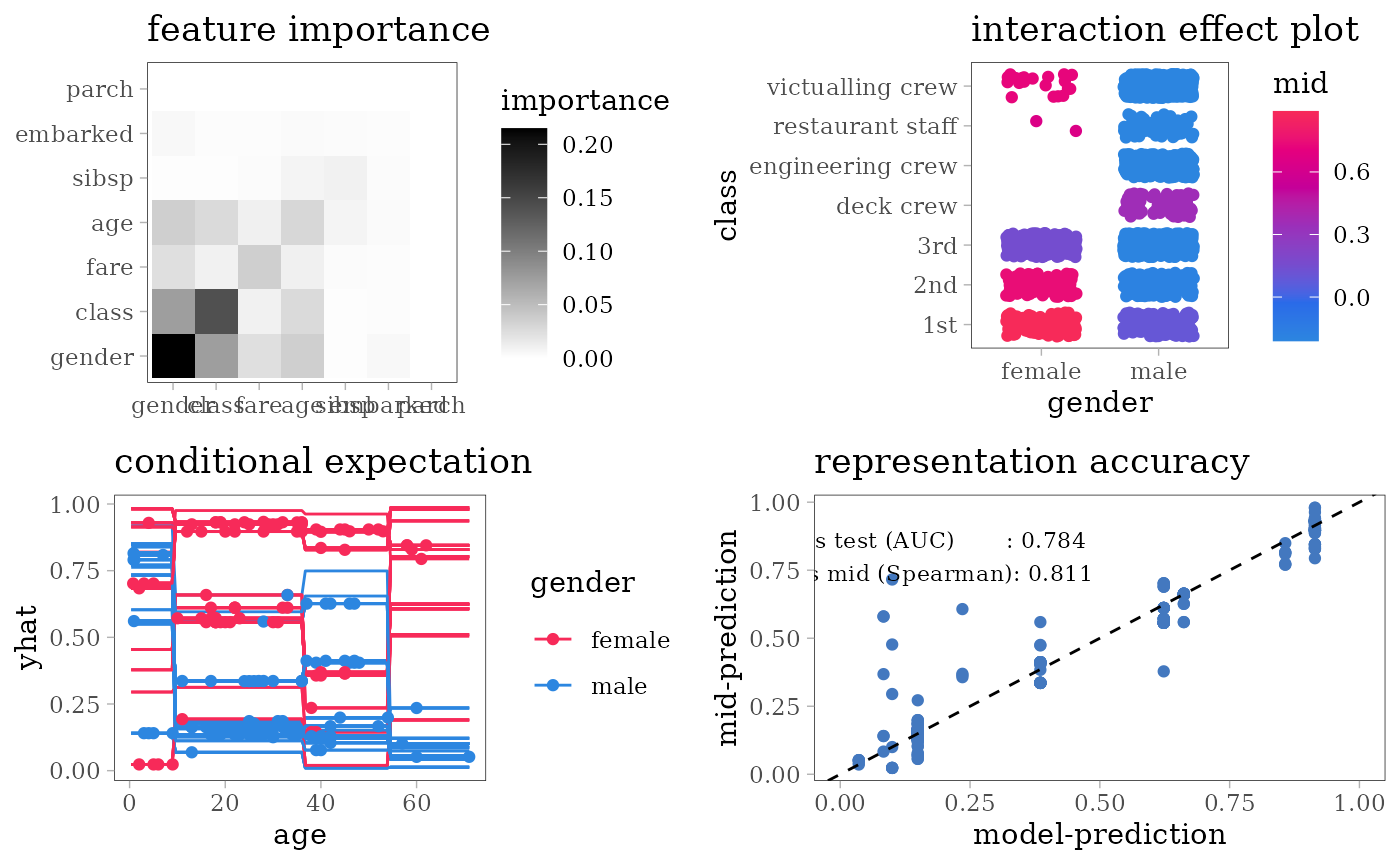
Other Models
Predictive MID
To fit a MID model for the Titanic classification task, we can use
“one-sided” link functions: identity-gaussian or
identity-logistic. These link functions map
to
and
to
,
while the inverse link functions map any real number to the value in the
unit interval
.
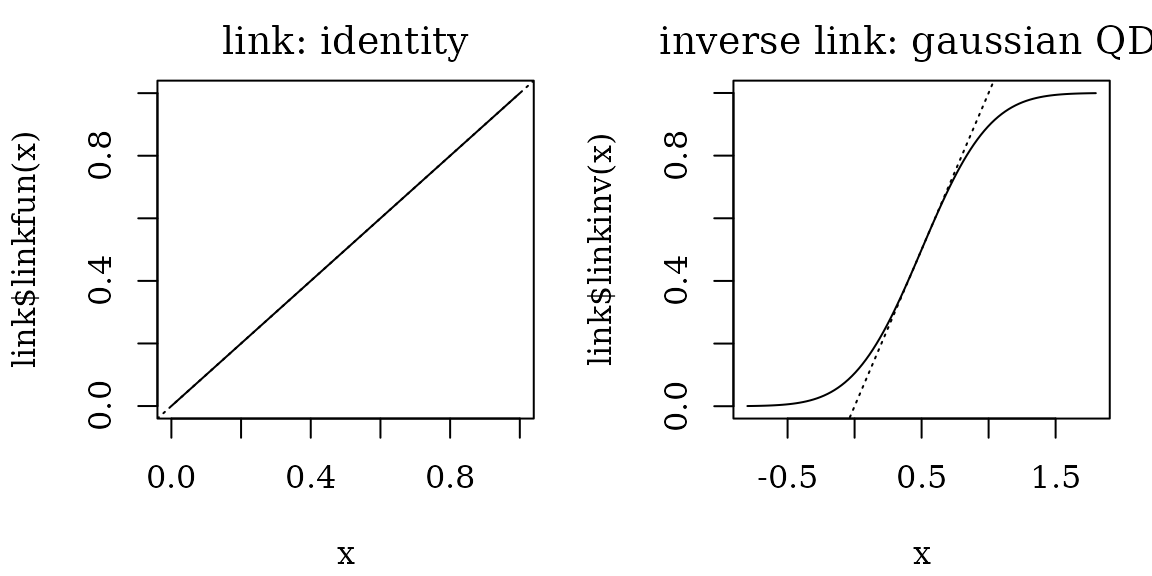
mid <- interpret(survived ~ .^2, train,
link = "identity-gaussian", lambda = .1)
#> 'model' not passed: response variable in 'data' is used
print(mid)
#>
#> Call:
#> interpret(formula = survived ~ .^2, data = train, link = "identity-gaussian",
#> lambda = 0.1)
#>
#> Intercept: 0.32285
#>
#> Main Effects:
#> 7 main effect terms
#>
#> Interactions:
#> 21 interaction terms
#>
#> Uninterpreted Variation Ratio: 0.61459
grid.arrange(grobs = effect_plots(mid), nrow = 2L)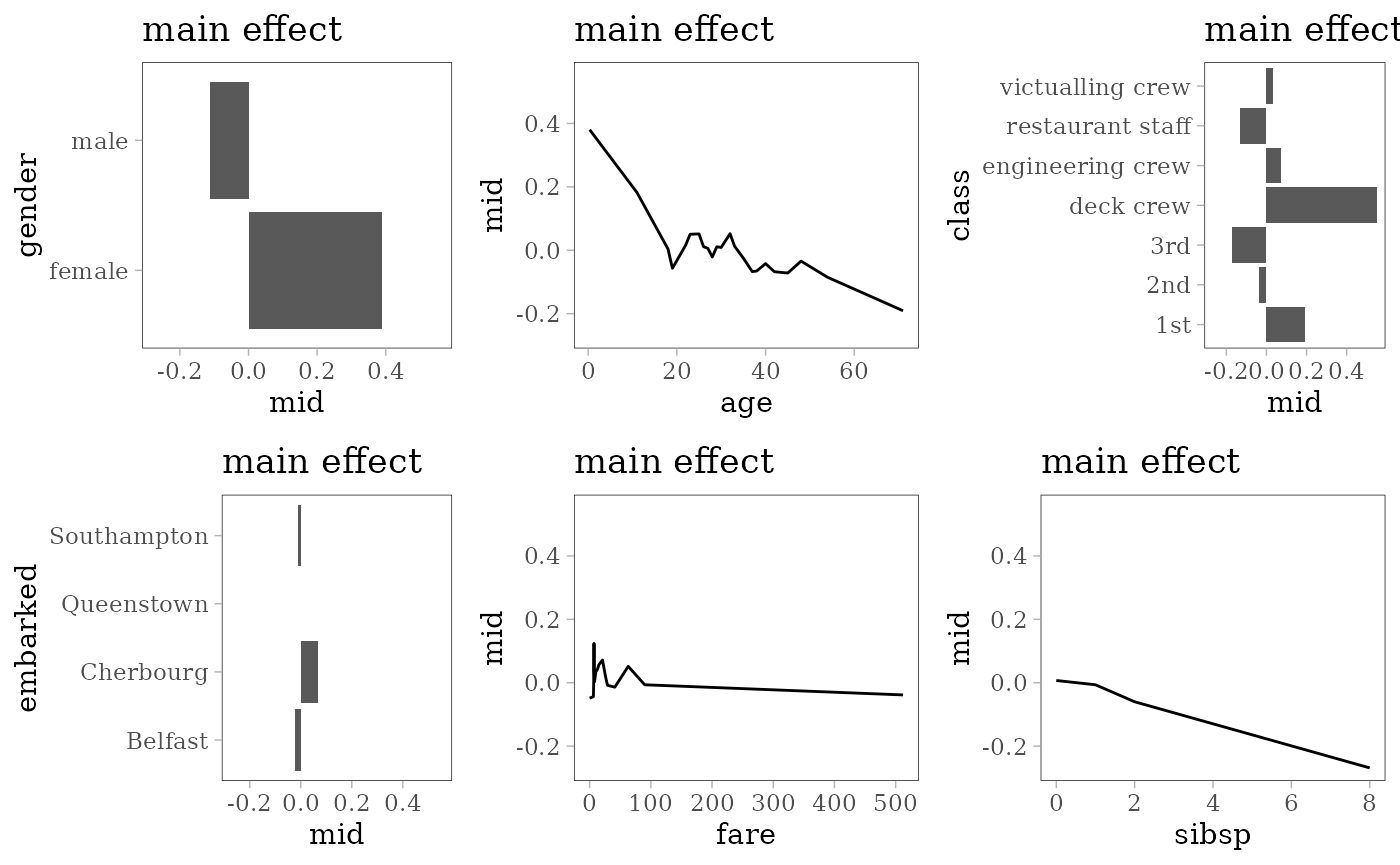
grid.arrange(nrow = 2L,
importance_plot(mid),
interaction_plot(mid),
ice_plot(mid),
interaction_plot(mid, "age:class"))